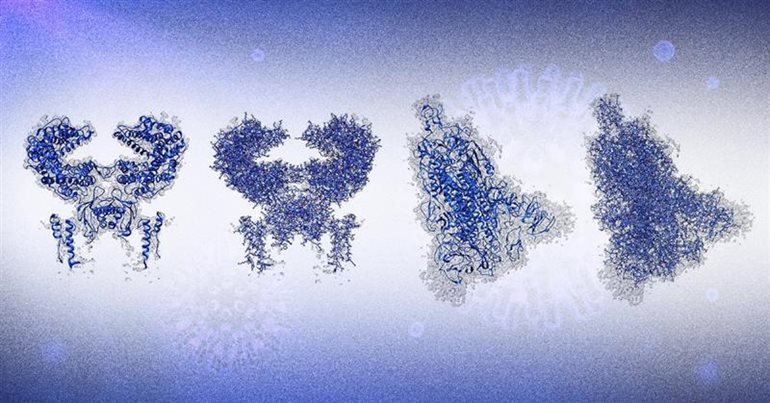
These are results produced by DeepTracer. The two at the left show the human receptor known as ACE2, to which the spike protein of the coronavirus binds. The two at the right show the coronavirus spike protein. The first and third images with the ribbon shapes represent the structures graphically, and the second and fourth images model the atomic structures in more detail.
By Douglas Esser
A team led by a computer scientist at the University of Washington Bothell is turning the power of artificial intelligence against the COVID-19 coronavirus with a new software tool that could help design vaccines.
Called DeepTracer, it uses deep learning to analyze a three-dimensional image of a virus protein molecule and trace the connections of its atoms.
“If you know the actual atomic structure of the viral protein, you will know how to speed up the development of vaccines or drugs,” said Dong Si, an assistant professor in the School of STEM’s Division of Computing & Software Systems.
Next-generation biomedicine

The virus images come from electron microscopes, recorded under super-cold cryogenic conditions. Cryogenic electron microscopy or cryo-EM was recognized with a 2017 Nobel prize. The method reduces electron irradiation damage to biological samples by freezing the specimens.
Different images can be combined for a 3D reconstruction. It’s called 3D electron microscopy or 3DEM. Still, the results don’t show everything scientists need to see. That’s why the images have to be further processed.
Si has worked in 3DEM for 11 years. Most researchers in the field use physics or traditional computational methods to predict atomic structure, Si said. He formed a data mining and deep learning research team a year ago to refine predictive models.
“We are one of the first groups in the world to use deep learning to do this three-dimensional EM atomic protein complex structure prediction,” he said.
Recognized impact
An independent expert in cryogenic electron microscopy looked at the potential of DeepTracer. Carlos Oscar Sánchez Sorzano, a group leader of the Biocomputing Unit of the National Center for Biotechnology / Instruct Image Processing Center in Madrid, Spain, tested DeepTracer and said it worked beautifully.
“[It fills] the gap between cryo-EM maps at high resolution and atomic modelling from sequence in a useful way that we hope opens new research lines leading to an even brighter future for the field,” Sorzano said. “The impact of this new algorithm goes well beyond the SARS-CoV-2 proteins and is widely applicable to any cryo-EM map below 3.5 angstroms, which are relatively common at present.” (An angstrom is one hundred-millionth of a centimeter.)
Available online
The DeepTracer website launched July 21, 2020. A scientific manuscript on the project posted the same day. Fully automatic and available to anyone in the world, DeepTracer drew a surge of interest. There were 1,264 visits and 277 new users from 20 countries the first day, Si said.
Users can upload their own 3DEM image or run data from an international collaboration called EMDataResource. It contains 3D images of macromolecular complexes, including the COVID-19 novel coronavirus as well as others such as MERS-CoV and SARS-CoV.
DeepTracer processes the image with an algorithm along with protein sequence information to predict where every atom is located. Compared to earlier state-of-the-art 3DEM modeling methods, results are more accurate and more complete.
“We can determine the atomic-level structure of a very large protein complex,” Si said.
The results, he said, are also faster — within a few minutes or hours, depending on the image size.
Making it happen
Si teaches courses in machine learning and artificial intelligence. His research team DAIS, the Data Analysis & Intelligent Systems group, applies those techniques to biomedical and social science problems. A previous project, for example, created a chatbot to help isolated caregivers deal with stress.
His cryo-EM deep learning project received a $200,000 National Science Foundation Rapid Response Research grant in May as part of an effort to mobilize the scientific community to respond to the coronavirus.
The team included 15 students, mostly from UW Bothell and the UW in Seattle, plus one high school student, Adrian Avram, from The Overlake School in Redmond, Washington, who emailed Si expressing interest in computer science and was invited to join.
Jonas Pfab, a graduate student in Computer Science & Software Engineering, is the lead engineer. Much of the front-end website and database engineering was done by Yinrui “Bobby” Deng, an undergraduate at the UW in Seattle majoring in Computational Economics and in Communication. Nhut Minh (Jack) Phan, a graduate student in Computer Science & Software Engineering, helped refine the algorithm and write the manuscript.
Among the next steps for the team is further improving the accuracy and efficiency of DeepTracer.
“Every day, I wake up in the morning and need to work on this project to make it faster, better and more accurate,” Si said. “I have a huge passion and dedication to it.”



